2. Use case II: Fusion genes detection from multiple myeloma patient RNA-seq#
2.1. Background#
Clinical Applications of RNA-Seq in Diagnostic Testing
RNA sequencing (RNA-Seq) is a high-throughput transcriptome profiling technology that enables comprehensive analysis of gene expression, splicing variants, fusion events, and novel transcripts. In clinical diagnostics, it serves as a powerful tool for:
Cancer Subtyping: Identifying tumor-specific gene expression signatures, fusion genes (e.g., BCR-ABL1), and aberrant splicing events to guide targeted therapies.
Rare Disease Diagnosis: Detecting dysregulated pathways and aberrant expression in Mendelian disorders where DNA-based tests are inconclusive.
Infectious Disease Characterization: Profiling host-pathogen interactions and pathogen expression in complex infections.
Biomarker Discovery: Validating expression-based biomarkers for disease monitoring and treatment response.
Gene fusions, or chromosomal translocations, are among the most common classes of mutations observed in cancer. These events can contribute to oncogenesis either by generating chimeric transcripts—such as BCR::ABL and RUNX1::RUNX1T1—or by inducing the overexpression of oncogenes, such as IGH::CCND1. The RNA-seq analysis module in ClinDet integrates key functionalities including transcript quantification, gene fusion detection, immune repertoire profiling, and RNA variant calling. In this study, we employed the ClinDet RNA-seq module to perform gene expression quantification and structural variant analysis by reanalyzing transcriptomic data from 31 flow-sorted bone marrow plasma cell samples published by Jaime et al. As a case study, we focused on three multiple myeloma patients (CD1, MS3, and MF1), all of whom were reported to carry chromosomal rearrangements involving the IGH enhancer and partner genes.
2.2. Setup a project folder#
Note
Before starting the analysis, please ensure that you have set up the analysis environment using the build_conda_env.sh script.
Create a folder named project/MM_RNA in your home directory and activate the clindet conda environment.
mkdir -p ~/projects/MM_RNA
cd ~/projects/MM_RNA
conda activate clindet
2.3. Download data and setup a samplesheet.csv#
Download Multiple myeloma RNA-seq data from the SRA database using wget and prepare the sample information file, make sure fastq-dump are in in $PATH (if don’t install it first)
cd ~/projects/MM_RNA
mkdir -p data && cd data
## Methods one multiple myeloma RNA-seq data
wget -q -c -O A26.11 https://sra-pub-run-odp.s3.amazonaws.com/sra/SRR12099713/SRR12099713
wget -q -c -O A27.19 https://sra-pub-run-odp.s3.amazonaws.com/sra/SRR12099714/SRR12099714
wget -q -c -O A28.15 https://sra-pub-run-odp.s3.amazonaws.com/sra/SRR12099715/SRR12099715
fastq-dump --gzip -O ~/projects/MM_RNA/data --split-3 ./A26.11
fastq-dump --gzip -O ~/projects/MM_RNA/data --split-3 ./A27.19
fastq-dump --gzip -O ~/projects/MM_RNA/data --split-3 ./A28.15
Next, create a CSV file named pipe_rna.csv in the ~/projects/MM_RNA directory with the following content:
R1_file_path,R2_file_path,Sample_name,Project
~/projects/MM_RNA/data/A26.11_1.fastq.gz,~/projects/MM_RNA/data/A26.11_2.fastq.gz,MF1
~/projects/MM_RNA/data/A27.19_1.fastq.gz,~/projects/MM_RNA/data/A27.19_2.fastq.gz,MS3
~/projects/MM_RNA/data/A28.15_1.fastq.gz,~/projects/MM_RNA/data/A28.15_2.fastq.gz,CD1
2.4. Write an Snakemake file from template#
For this project, modify the sample sheet and create a new Snakemake file named snake_rna.smk (see below). Set the following parameters in the Snakemake file:
configfile (str): config file for softwares and resource parameters.
stage (list): analysis steps. avaiable options:
['RSEM','arriba','TRUST4','samlom','kallisto']
2.5. write Snakemake file#
For this project, we need change the samplesheet info and config.yaml path in the snake_rna.smk .
Tip
import pandas as pd
samples_info = pd.read_csv('./pipe_rna.csv',index_col='Sample_name')
unpaired_samples = samples_info.loc[pd.isna(samples_info['R2_file_path'])].index.tolist()
paired_samples = samples_info.loc[~pd.isna(samples_info['R1_file_path'])].index.tolist()
configfile: "/AbsoPath/of/clindet/folder/config/config.yaml"
stages = ['RSEM','arriba','TRUST4','samlom','kallisto']
caller_list = ['sentieon_anno_rnaedit','Mutect2_filter']
project = 'RNA'
genome_version = 'b37'
rna_res_list = [
##### for isoform expression RSEM ######
"{project}/{genome_version}/results/summary/RSEM/{sample}/{sample}.genes.results" if 'RSEM' in rna_stages else None,
##### kallisto
"{project}/{genome_version}/results/summary/kallisto/{sample}/abundance.tsv" if 'kallisto' in rna_stages else None,
##### salmon
"{project}/{genome_version}/results/summary/salmon/{sample}/quant.sf" if 'salmon' in rna_stages else None,
##### for Immu analysis #####
"{project}/{genome_version}/results/IG/TRUST4/{sample}_report.tsv" if 'TRUST4' in rna_stages else None,
##### for fusion gene detection #####
"{project}/{genome_version}/results/fusion/{sample}_arriba_fusion.tsv" if 'arriba' in rna_stages else None,
##### for isofox immu analysis #####
"{project}/{genome_version}/results/summary/isofox/{sample}/{sample}.sorted.bam" if 'isofox' in rna_stages else None,
#### mutation section #####
"{project}/{genome_version}/results/mut/maf/{sample}/merge/{sample}.maf"
]
rna_res_list = list(filter(None, rna_res_list))
rule all:
input:
## paired sample
expand(rna_res_list,
sample = paired_samples,
project = project,
genome_version = genome_version
)
##### Modules #####
include: "/AbsoPath/of/clindet/folder/workflow/RNA/Snakefile"
2.6. Run ClinDet#
There is two way you can run ClinDet
run on a local server
submit to HPC through slurm
2.6.1. Run on local node#
nohup snakemake -j 30 --printshellcmds -s snake_rna.smk \
--use-singularity --singularity-args "--bind /your/home/path:/your/home/path" \
--latency-wait 300 --use-conda >> rna.log
2.6.2. Submit to HPC use slurm#
we provide a slurm config.yaml under clindet/workflow/config_slurm folder.
nohup snakemake --profile /Absolute/Path/of/clindet/workflow/config_slurm \
-j 30 --printshellcmds -s snake_rna.smk --use-singularity \
--singularity-args "--bind /your/home/path:/your/home/path" \
--latency-wait 300 --use-conda >> rna.log
2.7. Results#
After successful execution, you will see the following directory structure. The fusion folder contains the fusion gene detection results, and the summary folder contains the gene expression quantification results.
2.7.1. Overview of outputs#
~/projects/MM_RNA/b37/results
├── fusion # Fusion Gene Detection Results
├── mapped # STAR mapping results
│ └── STAR
├── mut # RNA Mutation Detection Results
│ ├── dedup
│ ├── maf # annotated MAF file
│ ├── STAR
│ └── vcf
├── IG # Immune repertoire reconstruction results
│ └── TRUST4
│
└── summary # Results of Gene Expression Quantification
├── kallisto
├── RSEM
└── salmon
2.7.2. arriba fusion genes#
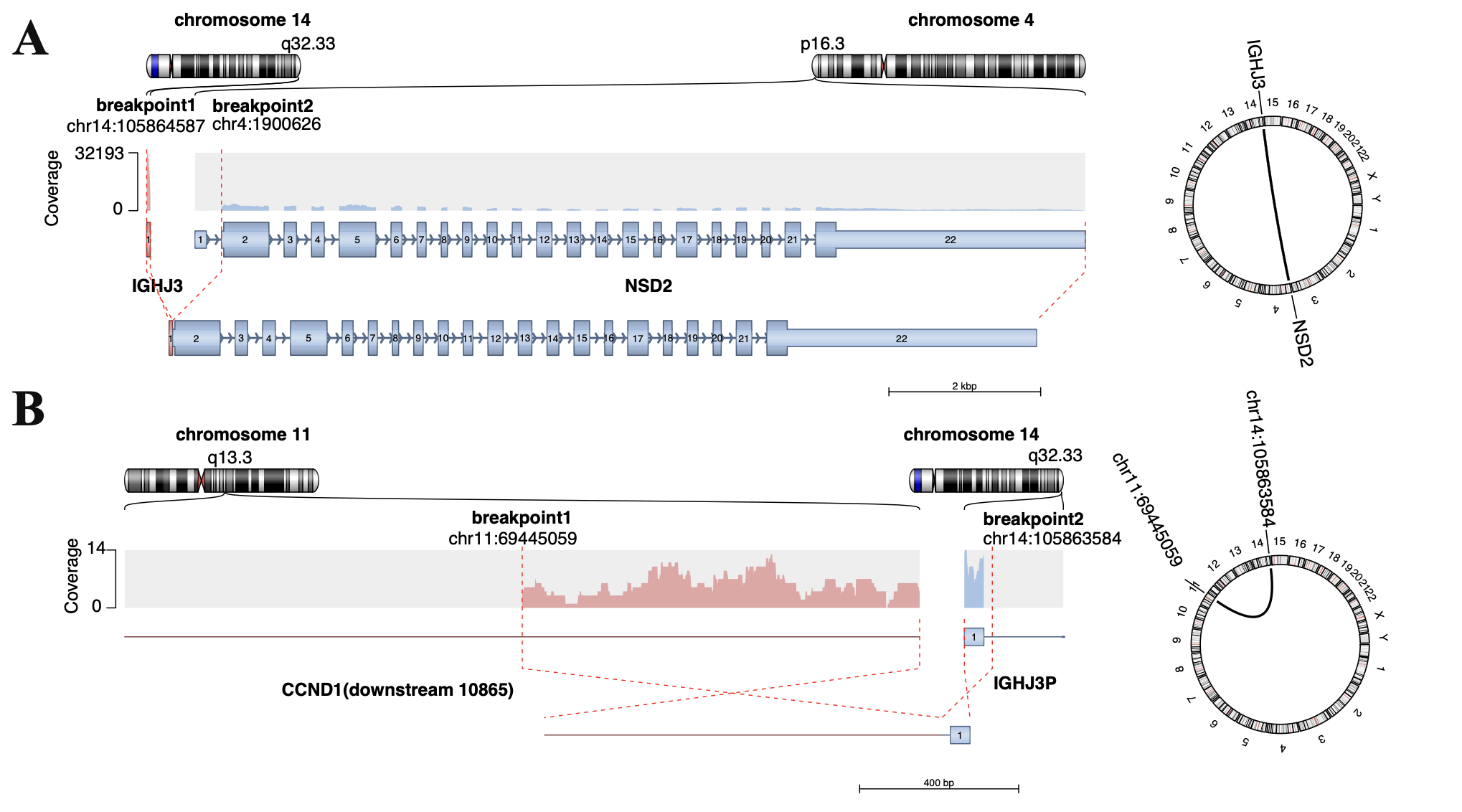
Within the gene fusion detection analysis, structural variants were identified in patients CD1 (IGH::CCND1) and MS3 (IGH::NSD2). In contrast, no detectable fusion transcript was found in patient MF1. However, all three patients exhibited aberrantly high expression levels of the corresponding partner genes. We hypothesize that the structural variation breakpoint in the IGH locus of patient MF1 may reside upstream of the MAF gene, possibly in a non-coding regulatory region, allowing enhancer-driven overexpression without producing a fusion transcript. IGH fusion genes circos plot (see below):
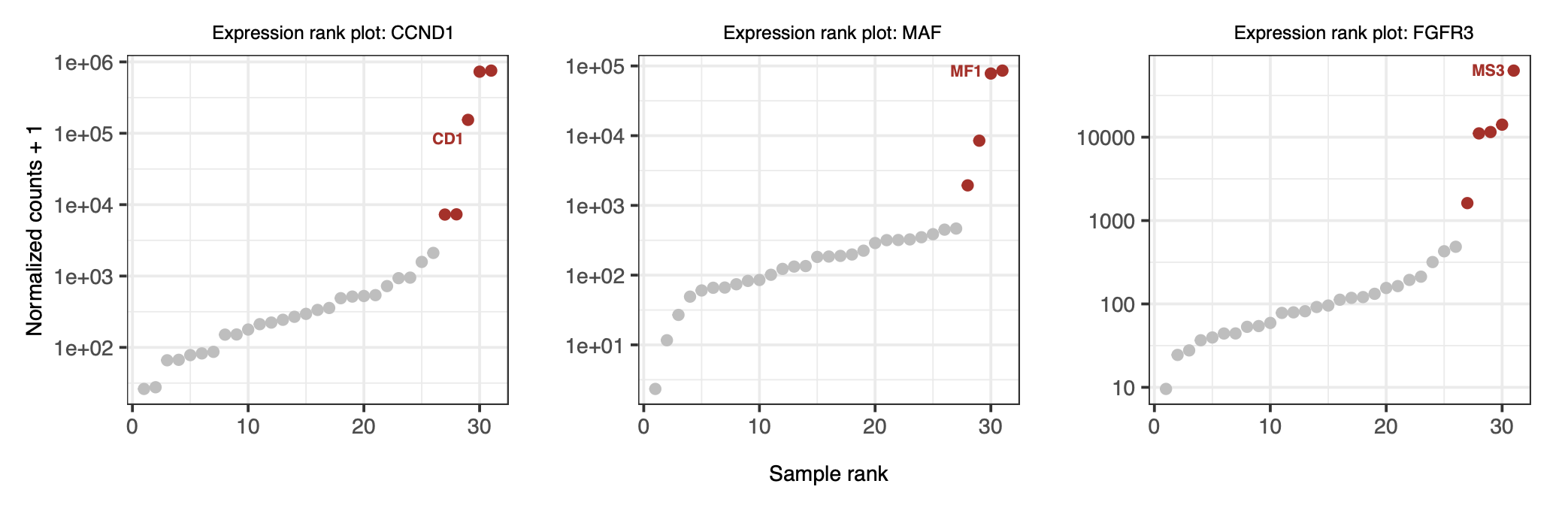
2.7.3. Aberrant expression of partner genes#
Additionally, if you wish to study gene expression levels, you can download the data for all samples from the original publication and perform quantification. Subsequently, use the Outliner package to analyze genes with aberrant expression within the patient cohort. In this example, we will not include this analysis, but interested readers are encouraged to download the data and explore it on their own, The figure below shows the expected analysis results.
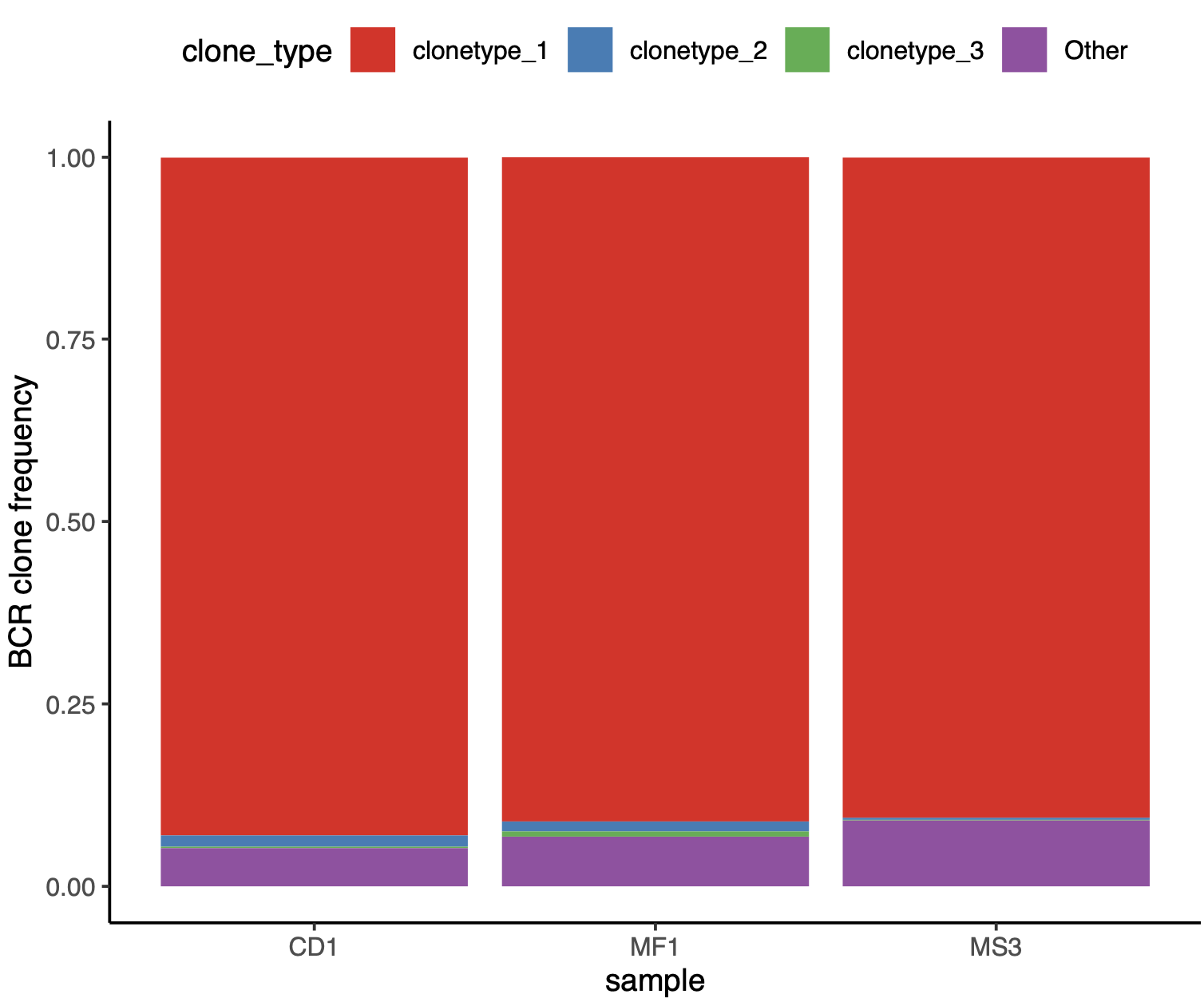
2.7.4. immune repertoire analysis#
Furthermore, immune repertoire profiling of the three samples revealed that more than 95% of the immunoglobulin sequences originated from a single clone. This finding is consistent with the prevailing hypothesis that multiple myeloma arises from a clonal expansion of a single progenitor B cell.