4. Use case IV: Quantifying the contributions of DNA repair defective gene mutations to mutational signatures(C.elegans)#
4.1. Background#
As mentioned by Douglas Hanahan in the “cancer hallmark paper (2022) (version 1: 2011)”, genome (DNA) instability and mutation is a fundamental component of cancer formation and pathogenesis. In the laset two decades, computational analysis of pan-cancer data has identified signatures of mutational processes thought to be responsible for the pattern of mutations in any given cancer. These analyses identified altered DNA repair pathways in a much broader spectrum of cancers than previously appreciated with significant therapeutic implications. The development of DNA repair deficiency biomarkers is critical to the implementation of therapeutic targeting of repair-deficient tumors, using either DNA damaging agents or immunotherapy for the personalization of cancer therapy (Jennifer Ma, et al. 2018). But the underly causal factor behind this process is hard to quantify in human, so as an alternative option, experiments have been done in model organisms, such as worm.
In this use case, we will re-analysis whole genome sequencing data from Volkova et,al. to validated the mutation signature pattern caused by gene mutations in DNA repaired pathway (e.g. xpc, mlh)
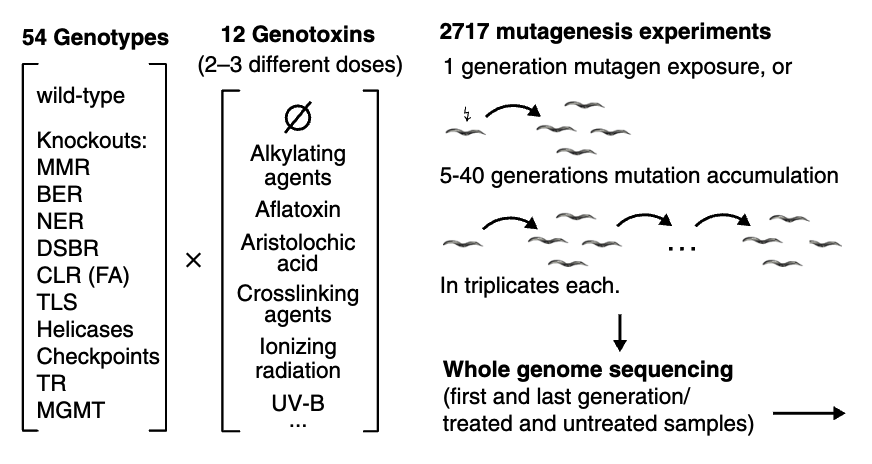
In the original paper, 54 gentypes C.elegans were treated by 12 genotoxins with 2-3 different doses, generated 2717 total mutagenesis experiments and whole genome sequencing data. We utilized Clindet’s tumor-normal paired mode within the WGS module to analyze this dataset and successfully reproduced the mutational signatures reported in the original study. Our focus was on WGS data from seven mutant worm samples and their matched normal controls, including: (1) three biological replicates of mlh-1 mutants, deficient in DNA mismatch repair and propagated for 20 generations; (2) three replicates of xpc-1 mutants exposed to UV light; and (3) one mrt-2 mutant, deficient in telomere maintenance and also propagated for 20 generations, which is known to undergo telomere crisis and breakage-fusion-bridge (BFB) cycles, resulting in extensive copy number alterations and genomic rearrangements.
here, we select ten samples which:
xpc-1 gene knockout with UV treat.
xpc-1 gene predicted to enable damaged DNA binding activity and single-stranded DNA binding activity. Involved in response to UV. Predicted to be located in nucleus. Predicted to be part of XPC complex and nucleotide-excision repair factor 2 complex. Predicted to be active in cytoplasm. Is expressed in germline precursor cell; intestine; and nervous system. Used to study xeroderma pigmentosum. Human ortholog(s) of this gene implicated in pancreatic cancer; serous cystadenocarcinoma; xeroderma pigmentosum; and xeroderma pigmentosum group C. Orthologous to human XPC (XPC complex subunit, DNA damage recognition and repair factor).
mlh-1 gene knockout.
mlh-1 gene predicted to enable ATP hydrolysis activity. Predicted to be involved in mismatch repair. Predicted to be located in nucleus. Predicted to be part of MutLalpha complex. Human ortholog(s) of this gene implicated in several diseases, including Lynch syndrome (multiple); carcinoma (multiple); and cervix uteri carcinoma in situ. Is an ortholog of human MLH1 (mutL homolog 1). Curator: Ranjana Kishore; Valerio Arnaboldi
mrt-2 gene knockout.
mrt-2 gene predicted to enable damaged DNA binding activity. Involved in DNA metabolic process and intracellular signal transduction. Predicted to be located in nucleus. Predicted to be part of checkpoint clamp complex. Orthologous to human RAD1 (RAD1 checkpoint DNA exonuclease).
Sample |
Genotype |
Generation |
Replicate |
Mutagen |
CD0009b |
mrt-2 |
0 |
0 |
|
CD0009f |
mrt-2 |
20 |
3 |
|
CD0001b |
N2 |
0 |
0 |
|
CD0134a |
mlh-1 |
20 |
2 |
|
CD0134c |
mlh-1 |
20 |
3 |
|
CD0134d |
mlh-1 |
20 |
4 |
|
CD0392a |
xpc-1 |
0 |
0 |
|
CD0842b |
xpc-1 |
1 |
1 |
UV |
CD0842c |
xpc-1 |
1 |
2 |
UV |
CD0842d |
xpc-1 |
1 |
3 |
UV |
4.2. Download data#
First, Create a folder named project/worm_WGS in your home directory and activate the Clindet conda environment.
mkdir -p ~/projects/worm_WGS
cd ~/projects/worm_WGS
conda activate clindet
To facilitate data download, we provide a metadata file worm_meta.tsv compiled from the original datasets. Please download this file and place it in the ~/projects/worm_WGS/ data directory. Afterwards, you can download the corresponding FASTQ files by using parallel (~ 35GB):
cd ~/projects/worm_WGS
mkdir -p data && cd data
parallel --colsep '\t' wget -q -c -O {12} {10} :::: worm_meta.tsv
4.4. Modify YAML file#
if you are not familiarity with yaml format, see ((en)[https:/yaml.org/], (zh)[https:/www.runoob.com/w3cnote/yaml-intro.html])
Users should configure the C.elegans genome settings in the config.yamlfile and ensure all referenced files are downloaded before beginning the analysis.
4.4.1. modify config.yaml for mutation detection#
Add WBcel235 config files to
config['resource']section
WBcel235:
REFFA: "/AbsoPath/of/clindet/folder/resources/ref_genome/WBcel235/WBcel235_genome.fa"
GTF: "/AbsoPath/of/clindet/folder/resources/ref_genome/WBcel235/Caenorhabditis_elegans.WBcel235.114.gtf"
GENOME_BED: ""
DBSNP: ""
DBSNP_GZ: "/AbsoPath/of/clindet/folder/reference/WBcel235/c_elegans.dbsnp.vcf.gz"
DBSNP_INDEL: "/AbsoPath/of/clindet/folder/reference/WBcel235/worm_fake_dbsnp.vcf.gz"
MUTECT2_VCF: "/AbsoPath/of/clindet/folder/reference/WBcel235/c_elegans.dbsnp.vcf.gz"
WGS_PON: ""
REFFA_DICT: "/AbsoPath/of/clindet/folder/resources/ref_genome/WBcel235/WBcel235_genome.dict"
add WBcel235 config
config['softwares']['vcf2maf']['build_version']andconfig['softwares']['vcf2maf']['vep']section
build_version:
WBcel235: "WBcel235"
vep:
WBcel235:
vep_data: "/AbsoPath/of/clindet/folder/resources/ref_genome/WBcel235/vep"
vep_path: "/Your/Path/of/vep/bin"
cache_version: "113"
species: "homo_sapiens"
Very Important: To ensure the vcf2maf tool can properly execute and complete variant annotation, users need to modify the code at line 466 in the vcf2maf.pl script (AbsoPath of conda env clindet_vep /bin/vcf2maf.pl).
Original code:
$vep_cmd .= " --buffer_size $buffer_size --sift b --ccds";
Modified code
# $vep_cmd .= " --no_stats --buffer_size $buffer_size --sift b --ccds";
$vep_cmd .= " --no_stats --buffer_size $buffer_size --ccds";
# set sift to false if not human or mouse data
$vep_cmd .= ( $species ~~ ['homo_sapiens','mus_musculus'] ? " --sift b" : " " );
4.5. write Snakemake file#
For this project, we need change the sample sheet info and setup a new snakemake file.
Create a CSV file named pipe_wes.csv in the ~/projects/worm_WGS directory with the following content:
Tumor_R1_file_path,Tumor_R2_file_path,Normal_R1_file_path,Normal_R2_file_path,Sample_name,Target_file_bed,Project
/AbsoPath/of/projects/worm_WGS/data/CD0009f_R1.fq.gz,/AbsoPath/of/projects/worm_WGS/data/CD0009f_R2.fq.gz,/AbsoPath/of/projects/worm_WGS/data/CD0009b-1_R1.fq.gz,/AbsoPath/of/projects/worm_WGS/data/CD0009b-1_R2.fq.gz,mrt-2,,C_elegans
/AbsoPath/of/projects/worm_WGS/data/CD0842b_R1.fq.gz,/AbsoPath/of/projects/worm_WGS/data/CD0842b_R2.fq.gz,/AbsoPath/of/projects/worm_WGS/data/CD0392a_R1.fq.gz,/AbsoPath/of/projects/worm_WGS/data/CD0392a_R2.fq.gz,CD0842b,,C_elegans
/AbsoPath/of/projects/worm_WGS/data/CD0842c_R1.fq.gz,/AbsoPath/of/projects/worm_WGS/data/CD0842c_R2.fq.gz,/AbsoPath/of/projects/worm_WGS/data/CD0392a_R1.fq.gz,/AbsoPath/of/projects/worm_WGS/data/CD0392a_R2.fq.gz,CD0842c,,C_elegans
/AbsoPath/of/projects/worm_WGS/data/CD0842d_R1.fq.gz,/AbsoPath/of/projects/worm_WGS/data/CD0842d_R2.fq.gz,/AbsoPath/of/projects/worm_WGS/data/CD0392a_R1.fq.gz,/AbsoPath/of/projects/worm_WGS/data/CD0392a_R2.fq.gz,CD0842d,,C_elegans
/AbsoPath/of/projects/worm_WGS/data/CD0134a_R1.fq.gz,/AbsoPath/of/projects/worm_WGS/data/CD0134a_R2.fq.gz,/AbsoPath/of/projects/worm_WGS/data/CD0097a_R1.fq.gz,/AbsoPath/of/projects/worm_WGS/data/CD0097a_R2.fq.gz,CD0134a,,C_elegans
/AbsoPath/of/projects/worm_WGS/data/CD0134c_R1.fq.gz,/AbsoPath/of/projects/worm_WGS/data/CD0134c_R2.fq.gz,/AbsoPath/of/projects/worm_WGS/data/CD0097a_R1.fq.gz,/AbsoPath/of/projects/worm_WGS/data/CD0097a_R2.fq.gz,CD0134c,,C_elegans
/AbsoPath/of/projects/worm_WGS/data/CD0134d_R1.fq.gz,/AbsoPath/of/projects/worm_WGS/data/CD0134d_R2.fq.gz,/AbsoPath/of/projects/worm_WGS/data/CD0097a_R1.fq.gz,/AbsoPath/of/projects/worm_WGS/data/CD0097a_R2.fq.gz,CD0134d,,C_elegans
create a snakemake file
snake_wgs_worm.smkfrom the template (bellow):
Tip
import pandas as pd
samples_info = pd.read_csv('/AbsoPath/of/pipe_WGS_worm.csv',index_col='Sample_name') # you need change this
unpaired_samples = samples_info.loc[pd.isna(samples_info['Normal_R1_file_path'])].index.tolist()
paired_samples = samples_info.loc[~pd.isna(samples_info['Normal_R1_file_path'])].index.tolist()
configfile: "/public/ClinicalExam/lj_sih/projects/project_clindet/build_log/config.yaml"
# project = samples_info["Project"].unique().tolist()[0]
project = 'test' # Users can either specify the projectparameter or use the first value from the projectcolumn in the samplesheet.csvfile. The result files will be saved in the corresponding project directory.
genome_version = 'WBcel235' # FASTQ files will be aligned to the genome version specified in the config.yamlfile to ensure consistency with the configuration.
import os
caller_list = ['HaplotypeCaller','strelkasomaticmanta','caveman','muse','cgppindel_filter','varscan2']
stages = [] # Non-human data did not support sample pairing testing and patient report generation
germ_caller_list = ['strelkamanta','caveman']
somatic_cnv_list = ['sequenza']
somatic_sv_list = ['svaba','gridss','delly','Manta']
### tumor only call
tumor_only_caller = []
tumor_only_cnv_caller = []
recall_pon = False
pre_pon_db = True
recall_pon_pindel = False
recal = False
include: "/AbsoPath/of/clindet/folder/workflow/WGS/Snakefile" # the ABSOLUTE path of clindet workflow WGS subfolder snakefile
## setup tools to run, based on genome version
paired_res_list = [
##### for QC report ######
# rules.conpair_contamination.output if 'conpair' in stages else None,
'{project}/{genome_version}/logs/paired/conpair/{sample}.done' if 'conpair' in stages else None,
##### for SNV/INDEL calling #####
"{project}/{genome_version}/results/maf/paired/{sample}/merge/{sample}.maf",
##### for CNV result ##### There is a bug for snakemake rules namelist when include *smk for 3-4 levels
# rules.paired_purple.output.qc if 'purple' in somatic_cnv_list else None, # purple call
"{project}/{genome_version}/results/cnv/paired/purple/{sample}/purple/{sample}.purple.qc" if 'purple' in somatic_cnv_list else None, # purple call
# rules.CNA_ASCAT.output.rdata if 'ASCAT' in somatic_cnv_list else None, # ASCAT call
"{project}/{genome_version}/results/cnv/paired/ascat/{sample}/{sample}_ASCAT.rdata" if 'ASCAT' in somatic_cnv_list else None, # ASCAT call
# rules.facets.output.qc if 'facets' in somatic_cnv_list else None, # facets call
"{project}/{genome_version}/results/cnv/paired/freec/{sample}/{sample}_config_freec.ini" if 'freec' in somatic_cnv_list else None, # Control-FREEC call
# rules.CNA_exomedepth.output.tsv if 'exomedepth' in somatic_cnv_list else None, # sequenza call
"{project}/{genome_version}/results/cnv/paired/exomedepth/{sample}/{sample}_exomedepth.tsv" if 'exomedepth' in somatic_cnv_list else None, # sequenza call
# rules.sequenza_call.output.segment if 'sequenza' in somatic_cnv_list else None, # sequenza call
"{project}/{genome_version}/results/cnv/paired/sequenza/{sample}/{sample}_segments.txt" if 'sequenza' in somatic_cnv_list else None, # sequenza call
##### for SV result #####
# somatic_sv_list = ['BRASS','delly','gridss','igcaller','linx','svaba','Manta']
# BRASS call
"{project}/{genome_version}/results/sv/paired/BRASS/{sample}/{sample}_brass.log" if 'BRASS' in somatic_sv_list else None, # purple call
# DELLY call
"{project}/{genome_version}/results/sv/paired/DELLY/{sample}/SV_delly_{sample}.vcf" if 'delly' in somatic_sv_list else None, # ASCAT call
# gridss call
"{project}/{genome_version}/results/sv/paired/gridss/{sample}/high_confidence_somatic.vcf.bgz" if 'gridss' in somatic_sv_list else None, # Control-FREEC call
# linx call
"{project}/{genome_version}/results/sv/paired/linx/{sample}/{sample}.linx.svs.tsv" if 'linx' in somatic_sv_list else None, # sequenza call
# svaba call
"{project}/{genome_version}/results/sv/paired/svaba/{sample}/{sample}.svaba.somatic.sv.vcf" if 'svaba' in somatic_sv_list else None, # sequenza call
# igcaller call
"{project}/{genome_version}/results/sv/paired/igcaller/{sample}/{sample}-T_IgCaller/{sample}-T_output_filtered.tsv" if 'igcaller' in somatic_sv_list else None, # sequenza call
# Manta call
"{project}/{genome_version}/results/vcf/paired/{sample}/Manta/results/variants/somaticSV.vcf.gz" if 'Manta' in somatic_sv_list else None, # sequenza call
#### Case report #####
'{project}/{genome_version}/results/report/{sample}/{sample}_cancer_report.html' if 'case_report' in stages else None,
#### Multiple QC report #####
'{project}/{genome_version}/results/multiqc_report.html' if 'multiqc' in stages else None,
]
paired_res_list = list(filter(None, paired_res_list))
## unpaired sample list
unpaired_res_list = [
##### for SNV/INDEL calling #####
"{project}/{genome_version}/results/maf/unpaired/{sample}/merge/{sample}.maf",
##### for CNV result ##### There is a bug for snakemake rules namelist when include *smk for 3-4 levels
# rules.paired_purple.output.qc if 'purple' in somatic_cnv_list else None, # purple call
"{project}/{genome_version}/results/cnv/unpaired/purple/{sample}/purple/{sample}.purple.qc" if 'purple' in tumor_only_cnv_caller else None,
### if you want call CNV from use tumor-only WES data, take you own risk
"{project}/{genome_version}/results/cnv/unpaired/freec/{sample}/{sample}-T.bam_ratio.txt.png" if 'freec' in tumor_only_cnv_caller else None,
]
unpaired_res_list = list(filter(None, unpaired_res_list))
recal = False ## will not recal for non-human data
rule all:
input:
## paired sample
expand(paired_res_list,
project = project,
genome_version = genome_version,
sample = paired_samples,
group = groups,
caller = caller_list),
## unpaired sample
expand(unpaired_res_list,
project = project,
genome_version = genome_version,
sample = unpaired_samples,
caller = caller_list)
4.6. Run clindet#
nohup snakemake --profile workflow/config_slurm \
-j 30 --printshellcmds -s snake_wgs_worm.smk \
--use-singularity \
--singularity-args "--bind /you/homepath/:/you/homepath/" \
--latency-wait 300 --use-conda --conda-frontend conda -k >> worm.out &
4.7. Results#
4.7.1. Overview of output#
For the final output results, please refer to the corresponding section in Use Case One Use case I: SNV and CNV calling from Whole exome sequencing data. In the following section, we will present the analyzed results derived from these raw outputs.
4.7.2. Mutation calling and signature analysis#
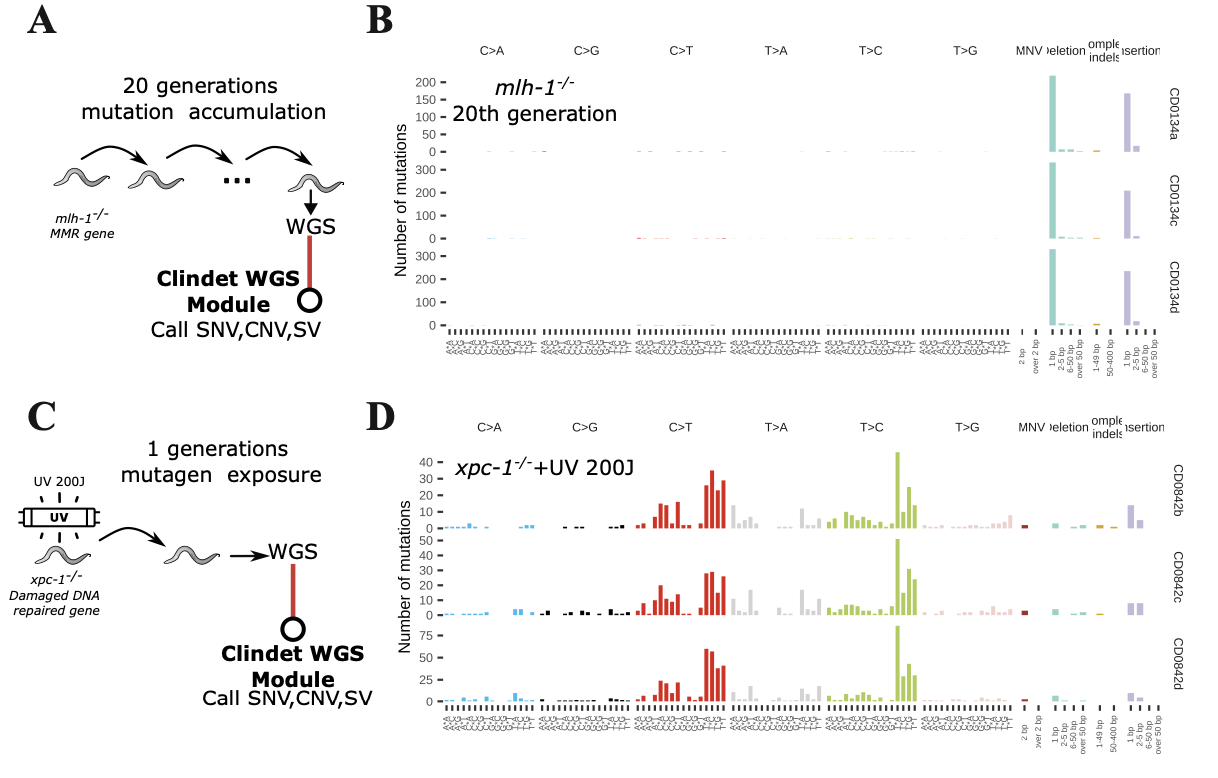
For somatic mutation detection, we employed multiple variant callers, including CaVEMan, MuSE, Pindel, Strelka2, and Manta. Copy number variations were identified using FragCounter, while structural variations were detected using Delly. Our analysis revealed that the mlh-1 mutant exhibited a mutational signature predominantly characterized by short insertions and deletions (indels), consistent with its deficiency in mismatch repair. The xpc-1 mutant, involved in nucleotide excision repair, showed an inability to correct UV-induced pyrimidine dimers, resulting in a mutational profile dominated by C>T (or T>C) transitions and short (1–5 bp) indels. Furthermore, in the mrt-2 mutant, Clindet identified CNVs and SVs localized to chromosome V, consistent with previously reported observations by Volkova et,al..
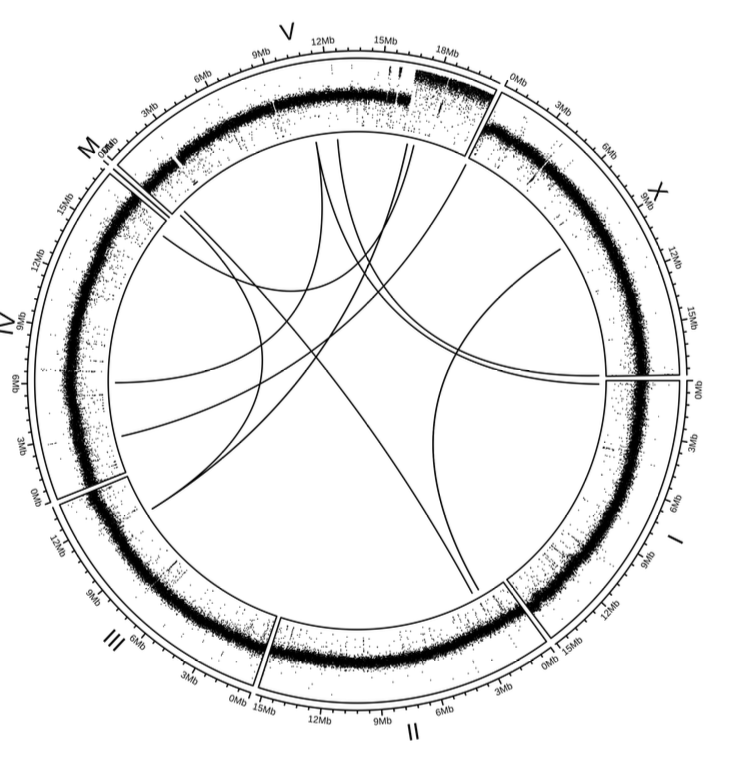
4.7.3. CNV and SV results#
Furthermore, in the mrt-2 mutant, Clindet identified CNVs and SVs localized to chromosome V, consistent with previously reported observations by Volkova et,al..
4.8. Optional: variant calling with CaVEMan and cgpindel#
When analyzing non-human sequencing data, tools like CaVEMan and Pindel require specific configurations to run successfully. However, some species may lack certain necessary data files, such as panel-of-normal results. As an alternative, a fake file can be provided to allow the pipeline to execute, but this may compromise the accuracy of variant calling results. Use this approach at your own risk.
These configuration files can be prepared according to the software documentation for CaVEMan and cgpPindel. In this example, we will use configurations based on the human b37 reference genome (ensure you have completed the setup from use case 1).
4.8.1. Setup a fake dbsnp.vcf file#
Users can create a dummy dbsnp.vcf file, compress it with bgzip (while retaining the original VCF file), and build an index for the compressed file using tabix. Then, replace the paths under the config[‘resources’][‘WBcel235’]section as follows:
Set both [‘DBSNP_GZ’]and [‘DBSNP_INDEL’]to the absolute path of the compressed file (e.g., dbsnp.vcf.gz).
Set [‘DBSNP’]to the path of the uncompressed VCF file (e.g., dbsnp.vcf).
Set [‘MUTECT2_VCF’]to the absolute path of the compressed file as well.
The specific content of this dummy VCF file should be structured as follows:
##fileformat=VCFv4.2 ##fileDate=20190602 ##contig=<ID=I,length=15072434> ##contig=<ID=II,length=15279421> ##contig=<ID=III,length=13783801> ##contig=<ID=IV,length=17493829> ##contig=<ID=V,length=20924180> ##contig=<ID=X,length=17718942> ##contig=<ID=MtDNA,length=13794> #CHROM POS ID REF ALT QUAL FILTER INFO
4.8.2. Modify config.yaml for CaVEMan and cgppindel#
In this example, we use the CaVEMan and cgppindel configuration for the human reference genome (b37). First, duplicate the content under the ['b37'] key within the config['singularity']['caveman'] section of the config.yaml file, and change the key name to WBcel235.
To enable the CaVEMan flagging process, you need to create a flag.vcf.config_Celegans.ini file in the directory /AbsoPath/of/clindet/folder/resources/ref_genome/b37/Sanger/SNV_INDEL_ref/caveman/flagging/ . Using flag.to.vcf.convert.ini file as a template, replace all instances of HUMAN_**with CELEGANS_**, and update the value of [‘flag’][‘c’] to the path of the flag.vcf.config_Celegans.ini file.
The final content under the config['singularity']['caveman'] and config['singularity']['cgppindel'] section should appear as shown below:
CaVEMan:
WBcel235:
ignorebed: "/AbsoPath/of/clindet/folder/reference/WBcel235/worm_ignore.bed"
flag:
c: "/AbsoPath/of/clindet/folder/reference/b37/SNV_INDEL_ref/caveman/flagging/flag.vcf.config_Celegans.ini"
v: "/AbsoPath/of/clindet/folder/reference/b37/SNV_INDEL_ref/caveman/flagging/flag.to.vcf.convert.ini"
u: "/AbsoPath/of/clindet/folder/reference/b37/SNV_INDEL_ref/caveman/flagging"
g: "/AbsoPath/of/clindet/folder/reference/b37/SNV_INDEL_ref/caveman/flagging/germline.bed.gz"
b: "/AbsoPath/of/clindet/folder/reference/b37/SNV_INDEL_ref/caveman/flagging"
ab: "/AbsoPath/of/clindet/folder/reference/b37/SNV_INDEL_ref/caveman/flagging"
s: "Celegans"
cgppindel:
WBcel235:
species: "C_elegans"
genes: "/AbsoPath/of/clindet/folder/resources/ref_genome/b37/Sanger/refarea_pindel/codingexon_regions.indel.bed.gz"
softfil: "/AbsoPath/of/clindet/folder/resources/ref_genome/b37/Sanger/SNV_INDEL_ref/pindel/WXS_Rules.lst"
simrep: "/AbsoPath/of/clindet/folder/resources/ref_genome/b37/Sanger/SNV_INDEL_ref/pindel/simpleRepeats.bed.gz"
normal_panel: "/AbsoPath/of/clindet/folder/resources/ref_genome/b37/Sanger/SNV_INDEL_ref/pindel/pindel_np.gff3.gz"
WES:
filter: "/AbsoPath/of/clindet/folder/resources/ref_genome/b37/Sanger/SNV_INDEL_ref/pindel/WXS_Rules.lst"
normal_panel: "/AbsoPath/of/clindet/folder/resources/ref_genome/b37/Sanger/SNV_INDEL_ref/pindel/pindel_np.gff3.gz"
WGS:
filter: "/AbsoPath/of/clindet/folder/resources/ref_genome/b37/Sanger/SNV_INDEL_ref/pindel/WGS_Rules.lst"
normal_panel: "/AbsoPath/of/clindet/folder/resources/ref_genome/b37/Sanger/SNV_INDEL_ref/pindel/pindel_np.gff3.gz"
