3. Use case III: Whole genome sequencing of COLO829 cell line#
3.1. Background#
Whole-genome sequencing (WGS) is increasingly being adopted in clinical oncology, providing a comprehensive view of the tumor genetic landscape. Compared to whole-exome sequencing (WES), this approach enables the detection of a broader range of disease-associated genetic alterations, including mutations in non-coding regions and structural variants (SVs). Such comprehensive profiling can facilitate deeper insights into cancer evolution and inform the development of personalized therapeutic strategies. However, WGS generates extensive datasets, necessitating robust bioinformatics pipelines for their analysis. To demonstrate how high-quality, validated somatic variants can be identified from WGS data, we employed Clindet to analyze a metastatic melanoma cell line (COLO829) and its matched normal lymphoblastoid cell line (COLO829BL), for which a “truth set” of copy number variants (CNVs) and structural variants (SVs) had been established.
COLO829 cell line
3.2. Setup a project folder#
Note
Before starting the analysis, please ensure that you have set up the analysis environment using the build_conda_env.sh script.
Create a folder named project/WGS in your home directory and activate the Clindet conda environment.
mkdir -p ~/projects/COLO829_WGS
cd ~/projects/COLO829_WGS
conda activate clindet
3.3. Download data and setup a samplesheet.csv#
Your can download WGS fastq file (~249G) from HMFtools Resources
cd ~/projects/COLO829_WGS
mkdir -p data && cd data
conda activate gsutil
gsutil -m cp \
"gs://hmf-public/HMFtools-Resources/test_data/COLO829v003T/fastq/COLO829v003R_AHHKYHDSXX_S13_L001_R1_001.fastq.gz" \
"gs://hmf-public/HMFtools-Resources/test_data/COLO829v003T/fastq/COLO829v003R_AHHKYHDSXX_S13_L001_R2_001.fastq.gz" \
"gs://hmf-public/HMFtools-Resources/test_data/COLO829v003T/fastq/COLO829v003R_AHHKYHDSXX_S13_L002_R1_001.fastq.gz" \
"gs://hmf-public/HMFtools-Resources/test_data/COLO829v003T/fastq/COLO829v003R_AHHKYHDSXX_S13_L002_R2_001.fastq.gz" \
"gs://hmf-public/HMFtools-Resources/test_data/COLO829v003T/fastq/COLO829v003R_AHHKYHDSXX_S13_L003_R1_001.fastq.gz" \
"gs://hmf-public/HMFtools-Resources/test_data/COLO829v003T/fastq/COLO829v003R_AHHKYHDSXX_S13_L003_R2_001.fastq.gz" \
"gs://hmf-public/HMFtools-Resources/test_data/COLO829v003T/fastq/COLO829v003R_AHHKYHDSXX_S13_L004_R1_001.fastq.gz" \
"gs://hmf-public/HMFtools-Resources/test_data/COLO829v003T/fastq/COLO829v003R_AHHKYHDSXX_S13_L004_R2_001.fastq.gz" \
"gs://hmf-public/HMFtools-Resources/test_data/COLO829v003T/fastq/COLO829v003T_AHHKYHDSXX_S12_L001_R1_001.fastq.gz" \
"gs://hmf-public/HMFtools-Resources/test_data/COLO829v003T/fastq/COLO829v003T_AHHKYHDSXX_S12_L001_R2_001.fastq.gz" \
"gs://hmf-public/HMFtools-Resources/test_data/COLO829v003T/fastq/COLO829v003T_AHHKYHDSXX_S12_L002_R1_001.fastq.gz" \
"gs://hmf-public/HMFtools-Resources/test_data/COLO829v003T/fastq/COLO829v003T_AHHKYHDSXX_S12_L002_R2_001.fastq.gz" \
"gs://hmf-public/HMFtools-Resources/test_data/COLO829v003T/fastq/COLO829v003T_AHHKYHDSXX_S12_L003_R1_001.fastq.gz" \
"gs://hmf-public/HMFtools-Resources/test_data/COLO829v003T/fastq/COLO829v003T_AHHKYHDSXX_S12_L003_R2_001.fastq.gz" \
"gs://hmf-public/HMFtools-Resources/test_data/COLO829v003T/fastq/COLO829v003T_AHHKYHDSXX_S12_L004_R1_001.fastq.gz" \
"gs://hmf-public/HMFtools-Resources/test_data/COLO829v003T/fastq/COLO829v003T_AHHKYHDSXX_S12_L004_R2_001.fastq.gz" \
.
then you need merge those fastq.gz files (eg. cat) to :
COLO829v003R_R1.fastq.gz (~26G)
COLO829v003R_R2.fastq.gz (~27G)
COLO829v003T_R1.fastq.gz (~96G)
COLO829v003T_R2.fastq.gz (~100G)
Next, create a CSV file named pipe_wgs.csv in the ~/projects/COLO829_WGS directory with the following content:
Tumor_R1_file_path,Tumor_R2_file_path,Normal_R1_file_path,Normal_R2_file_path,Sample_name,Project
/AbsoPath/of/projects/COLO829_WGS/data/COLO829v003T_R1.fastq.gz,/AbsoPath/of/projects/COLO829_WGS/data/COLO829v003T_R2.fastq.gz,/AbsoPath/of/projects/COLO829_WGS/data/COLO829v003R_R1.fastq.gz,/AbsoPath/of/projects/COLO829_WGS/data/COLO829v003R_R2.fastq.gz,COL0829,WGS
3.4. Write an Snakemake file from template#
For this project, modify the sample sheet and create a new Snakemake file named snake_wgs.smk (see below). Set the following parameters in the Snakemake file:
configfile (str): config file for softwares and resource parameters.
stage (list): analysis steps. avaiable options:
['conpair','report']sample_csv (str):
sample_info.csvpathgenome_version (str): genome version, can be genome version which setup in cofig.yaml eg.
b37recal (Boolean): use GATK BaseRecalibrator for Base Quality Score Recalibration? this is a time-consume task but can slightly improved calling accuracy.
True or False. defaultTrue.caller_list (list): somatic mutation calling softwares used. avaiable options:
['sage','HaplotypeCaller','strelkasomaticmanta','cgppindel_filter','caveman','muse','deepvariant','Mutect2_filter']germ_call_list (list): germline mutation calling softwares used. avaiable options:
['strelkamanta','caveman']somatic_cnv_list (list): Somatic Copy Number variant calling softwares. avaiable options:
['purple','ASCAT','sequenza','freec','exomedepth']recall_pon (Boolean): call panel-of-normal mutect2_pon.vcf from cohort normal samples. default
False, use public available resource.custome_pon_db (Boolean): use public available resource. default
False.recall_pon_pindel (Boolean): call panel-of-normal pindel_pon.vcf from cohort normal samples. default
False, use public available resource.
3.5. write Snakemake file#
For this project, we need change the sample sheet info.
Tip
import pandas as pd
samples_info = pd.read_csv('./pipe_WGS.csv',index_col='Sample_name')
unpaired_samples = samples_info.loc[pd.isna(samples_info['Normal_R1_file_path'])].index.tolist()
paired_samples = samples_info.loc[~pd.isna(samples_info['Normal_R1_file_path'])].index.tolist()
configfile: "/AbsoPath/of/clindet/folder/config/config.yaml"
project = samples_info["Project"].unique().tolist()[0]
genome_version = 'b37'
import os
germ_caller_list = ['caveman','deepvariant']
somatic_caller_list = ['strelkasomaticmanta','muse','cgppindel_filter','deepvariant','sage','caveman']
somatic_cnv_list = ['purple','ascat']
somatic_sv_list = ['svaba','gridss']
purple_sv = 'svaba'
recall_pon = False
custome_pon_db = True
recall_pon_pindel = False
paired_res_list = [
##### for QC report ######
# rules.conpair_contamination.output if 'conpair' in stages else None,
'{project}/{genome_version}/logs/paired/conpair/{sample}.done' if 'conpair' in stages else None,
##### for SNV/INDEL calling #####
"{project}/{genome_version}/results/maf/paired/{sample}/merge/{sample}.maf",
##### for CNV result ##### There is a bug for snakemake rules namelist when include *smk for 3-4 levels
# rules.paired_purple.output.qc if 'purple' in somatic_cnv_list else None, # purple call
"{project}/{genome_version}/results/cnv/paired/purple/{sample}/purple/{sample}.purple.qc" if 'purple' in somatic_cnv_list else None, # purple call
# rules.CNA_ASCAT.output.rdata if 'ASCAT' in somatic_cnv_list else None, # ASCAT call
"{project}/{genome_version}/results/cnv/paired/ascat/{sample}/{sample}_ASCAT.rdata" if 'ASCAT' in somatic_cnv_list else None, # ASCAT call
# rules.facets.output.qc if 'facets' in somatic_cnv_list else None, # facets call
# rules.facets.output.qc if 'freec' in somatic_cnv_list else None, # freec call
"{project}/{genome_version}/results/cnv/paired/freec/{sample}/{sample}_config_freec.ini" if 'freec' in somatic_cnv_list else None, # Control-FREEC call
# rules.CNA_exomedepth.output.tsv if 'exomedepth' in somatic_cnv_list else None, # sequenza call
"{project}/{genome_version}/results/cnv/paired/exomedepth/{sample}/{sample}_exomedepth.tsv" if 'exomedepth' in somatic_cnv_list else None, # sequenza call
# rules.sequenza_call.output.segment if 'sequenza' in somatic_cnv_list else None, # sequenza call
"{project}/{genome_version}/results/cnv/paired/sequenza/{sample}/{sample}_segments.txt" if 'sequenza' in somatic_cnv_list else None, # sequenza call
##### for SV result #####
# somatic_sv_list = ['BRASS','delly','gridss','igcaller','linx','svaba','Manta']
# BRASS call
"{project}/{genome_version}/results/sv/paired/BRASS/{sample}/{sample}_brass.log" if 'BRASS' in somatic_sv_list else None, # purple call
# DELLY call
"{project}/{genome_version}/results/sv/paired/DELLY/{sample}/SV_delly_{sample}.vcf" if 'delly' in somatic_sv_list else None, # ASCAT call
# gridss call
"{project}/{genome_version}/results/sv/paired/gridss/{sample}/high_confidence_somatic.vcf.bgz" if 'gridss' in somatic_sv_list else None, # Control-FREEC call
# linx call
"{project}/{genome_version}/results/sv/paired/linx/{sample}/{sample}.linx.svs.tsv" if 'linx' in somatic_sv_list else None, # sequenza call
# svaba call
"{project}/{genome_version}/results/sv/paired/svaba/{sample}/{sample}.svaba.somatic.sv.vcf" if 'svaba' in somatic_sv_list else None, # sequenza call
# igcaller call
"{project}/{genome_version}/results/sv/paired/igcaller/{sample}/{sample}-T_IgCaller/{sample}-T_output_filtered.tsv" if 'igcaller' in somatic_sv_list else None, # sequenza call
# Manta call
"{project}/{genome_version}/results/vcf/paired/{sample}/Manta/results/variants/somaticSV.vcf.gz" if 'Manta' in somatic_sv_list else None, # sequenza call
#### Case report #####
'{project}/{genome_version}/results/report/{sample}/{sample}_cancer_report.html' if 'case_report' in stages else None,
#### Multiple QC report #####
'{project}/{genome_version}/results/multiqc_report.html' if 'multiqc' in stages else None,
]
paired_res_list = list(filter(None, paired_res_list))
## unpaired sample list
unpaired_res_list = [
##### for SNV/INDEL calling #####
"{project}/{genome_version}/results/maf/unpaired/{sample}/merge/{sample}.maf",
##### for CNV result ##### There is a bug for snakemake rules namelist when include *smk for 3-4 levels
# rules.paired_purple.output.qc if 'purple' in somatic_cnv_list else None, # purple call
"{project}/{genome_version}/results/cnv/unpaired/purple/{sample}/purple/{sample}.purple.qc" if 'purple' in tumor_only_cnv_caller else None,
### if you want call CNV from use tumor-only WES data, take you own risk
"{project}/{genome_version}/results/cnv/unpaired/freec/{sample}/{sample}-T.bam_ratio.txt.png" if 'freec' in tumor_only_cnv_caller else None,
]
unpaired_res_list = list(filter(None, unpaired_res_list))
##### Modules #####
rule all:
input:
## paired sample
expand(paired_res_list,
sample = paired_samples,
project = project,
genome_version = genome_version,
group = groups,
caller = caller_list),
#### unpaired sample
expand(unpaired_res_list,
project = project,
genome_version = genome_version,
sample = unpaired_samples,
caller = caller_list),
##### multiqc report ########
f'{project}/{genome_version}/results/multiqc/filelist.txt' if 'report' in stages else [],
f'{project}/{genome_version}/results/multiqc_report.html' if 'report' in stages else [],
include: "workflow/WGS/Snakefile"
3.6. Run clindet#
There is two way you can run clindet
run on a local server
submit to HPC through slurm
3.6.1. Run on local node#
nohup snakemake -j 30 --printshellcmds -s snake_wgs.smk \
--use-singularity --singularity-args "--bind /your/home/path:/your/home/path" \
--latency-wait 300 --use-conda >> wgs.log
3.6.2. Submit to HPC use slurm#
we provide a slurm config.yaml under clindet/workflow/config_slurm folder.
nohup snakemake --profile workflow/config_slurm \
-j 30 --printshellcmds -s snake_wgs.smk --use-singularity \
--singularity-args "--bind /your/home/path:/your/home/path" \
--latency-wait 300 --use-conda >> wgs.log
3.7. Results#
After successful execution, you will see the following directory structure. The cnv folder contains the copy number variants detection results, the sv folder contains the structural variants detection results, and the vcf/maf folders contain mutaions (annotated and raw) results.
3.7.1. Overview of outputs#
~/projects/WGS/b37/results
├── cnv ** Copy Number variants results**
│ └── paired ** For tumor-normal paired sample**
│ ├── ascat
│ └── purple
├── dedup ** deduplication BAM files**
│ └── paired
├── maf ** annotation somatic mutation MAF files**
│ └── paired
│ └── COL0829
├── qc ** QC results for fastp conpair and so on**
│ └── dedup
│ └── paired
├── recal ** BAM files after base recalibration**
│ └── paired
├── sv ** Structural Variant results**
│ └── paired
│ ├── BRASS
│ ├── DELLY
│ ├── gridss
│ ├── linx
│ └── svaba
├── vcf
└── paired
├── COL0829
└── HG008
3.7.2. Copy number variants of COLO829#
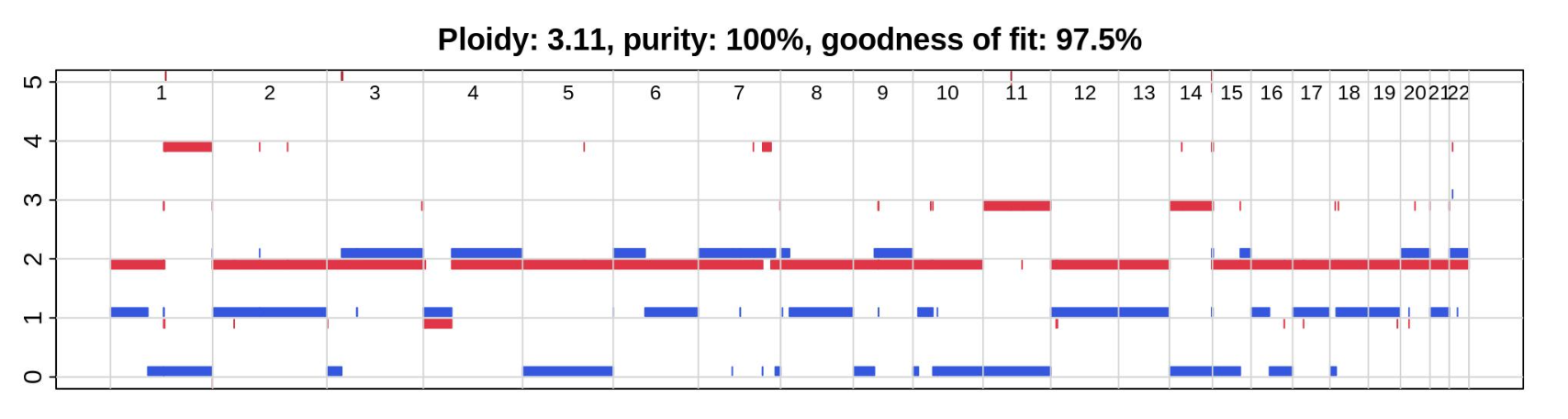
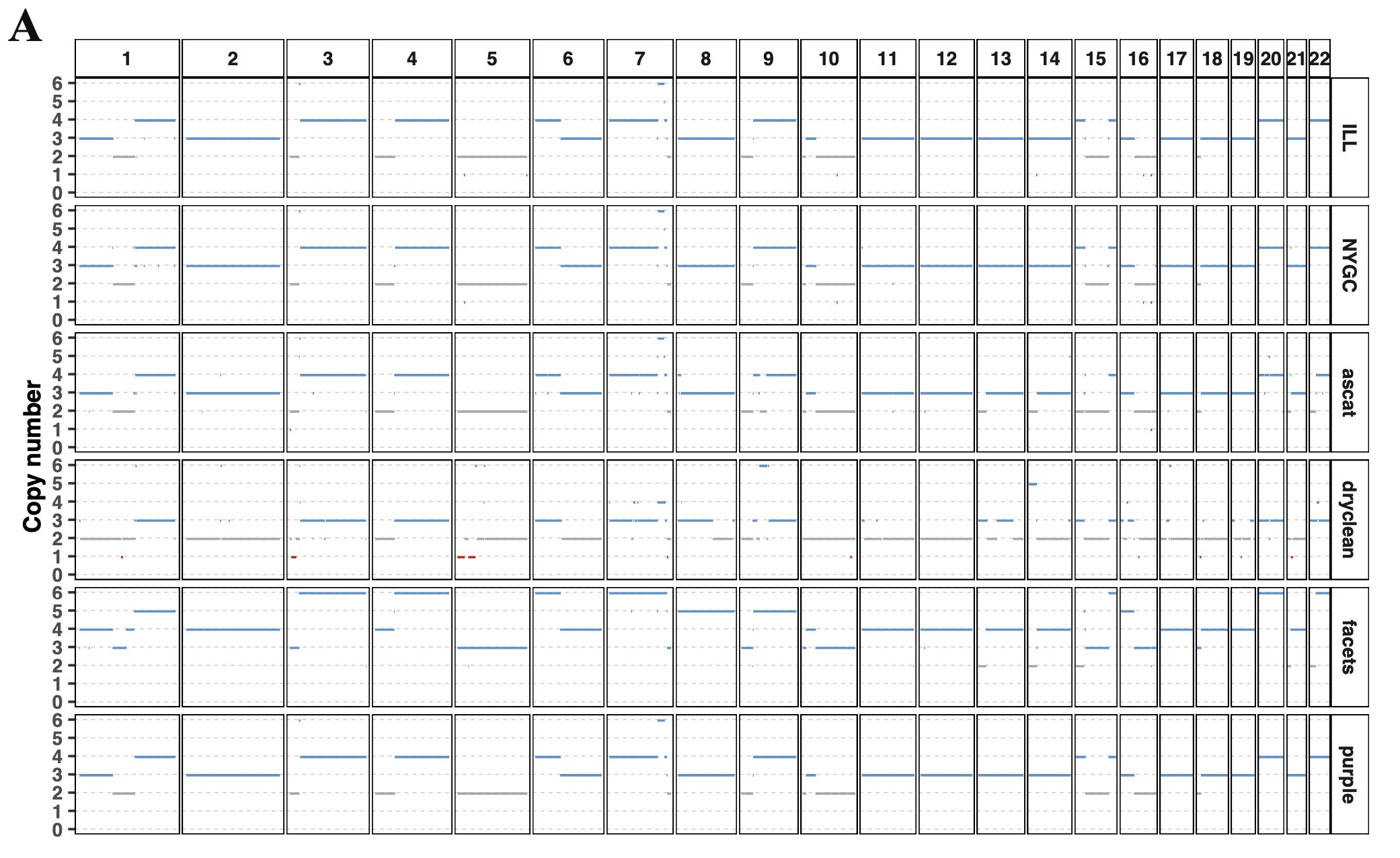
To assess the consistency among different software tools in representing the genomic content of the COLO829 cancer cell line, we evaluated the presence of CNVs and SVs. Regarding copy number variation, the four software tools generated similar estimates of tumor purity and ploidy.
The karyotype of the COLO829 cell line

Copy number results of COLO829
Furthermore, low-resolution copy number alteration (CNA) analysis revealed highly consistent copy number profiles across external “true set” software tools except for Facets, with correlation coefficients of 0.76–0.98 among different datasets.
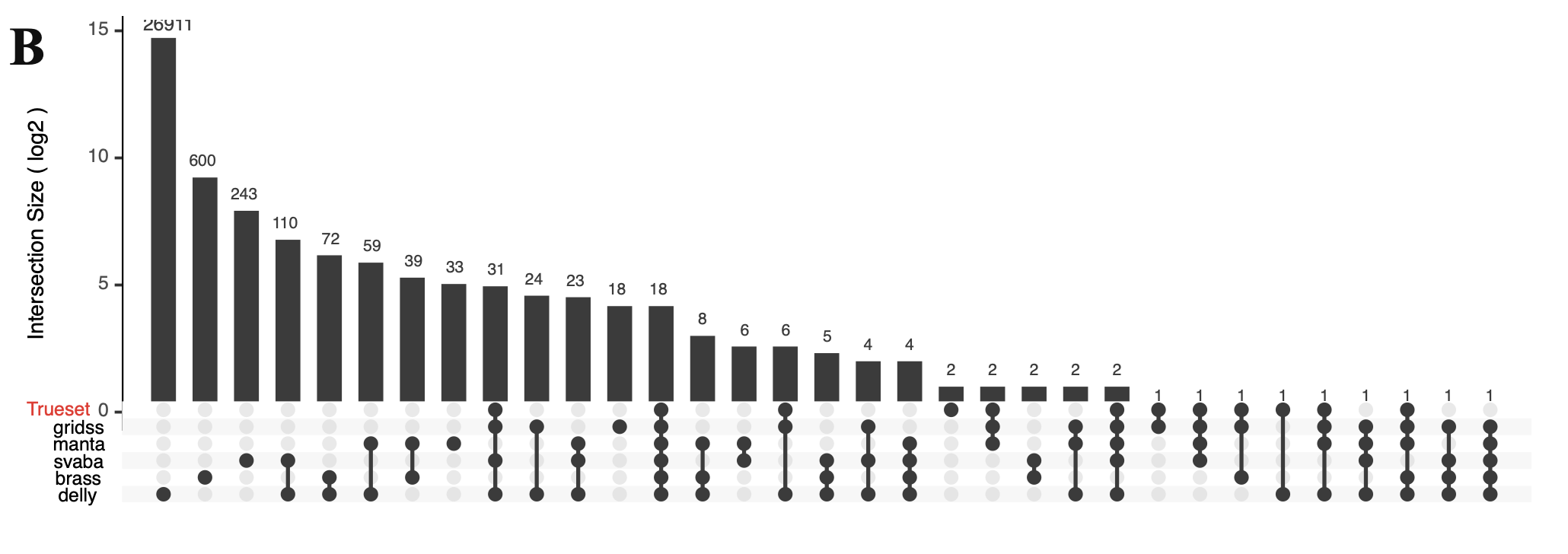
3.7.3. Structural variants of COLO829#
Due to the absence of established benchmarks and best-practice protocols for somatic SV detection, the primary aim of this study was to establish an analytical workflow rather than to benchmark SV calling tools. Consequently, we selected optimal mapping and SV calling tools based on current best practices and available knowledge. SV calling parameters were optimized for high sensitivity rather than maximum precision to minimize the risk of missing genuine events. Compared to the previously established “truth set” from XX et al., the candidate somatic SV calls produced by individual software tools showed considerable variability, ranging from 115 breakpoints detected by GRIDSS to 27,285 by DELLY, resulting in a combined total of 28,233 merged SV calls. Among these tools, DELLY generated a relatively high number of false-positive calls. The results indicated that integrating outputs from multiple SV detection tools substantially reduced the number of false-positive predictions.